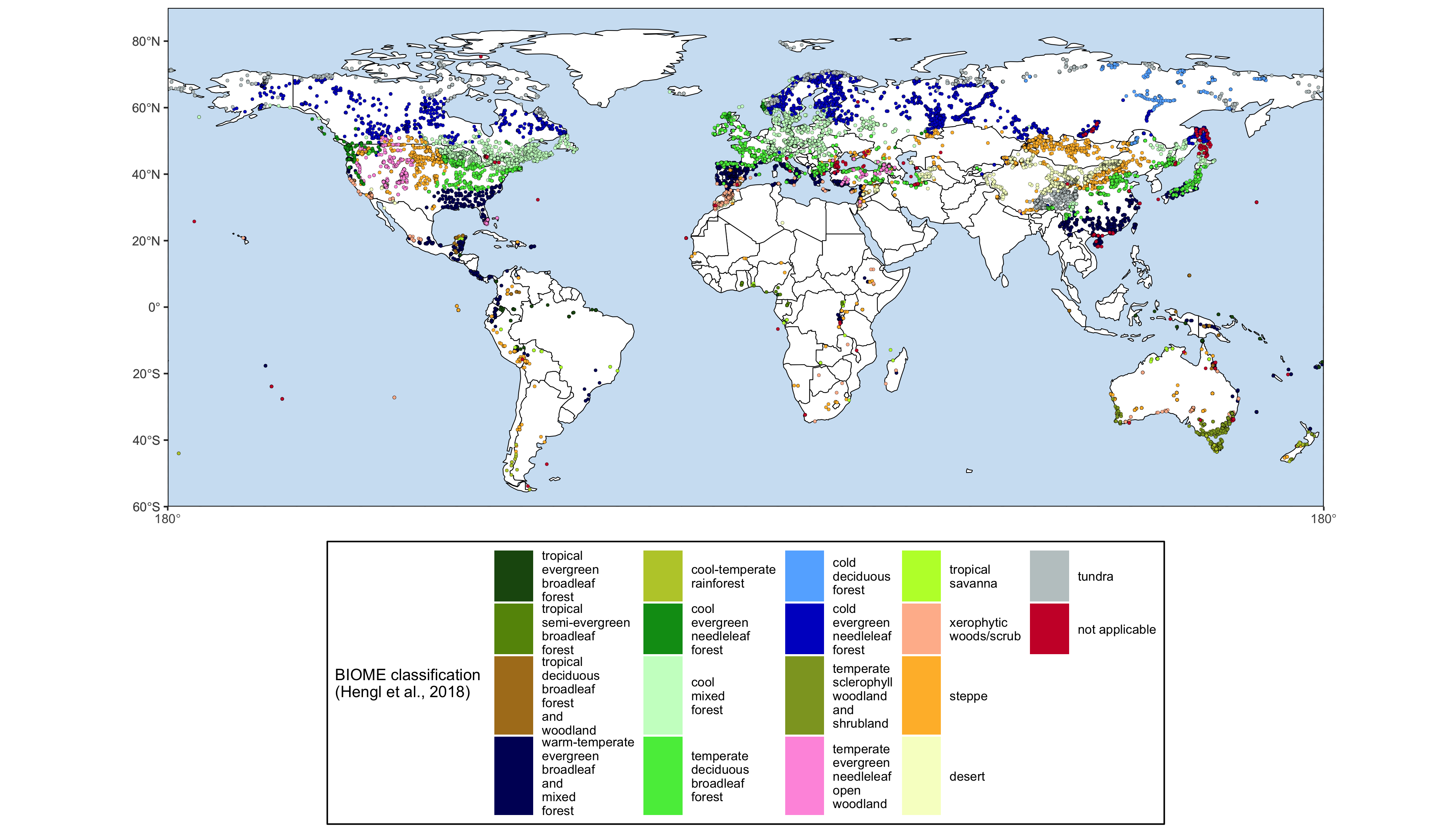
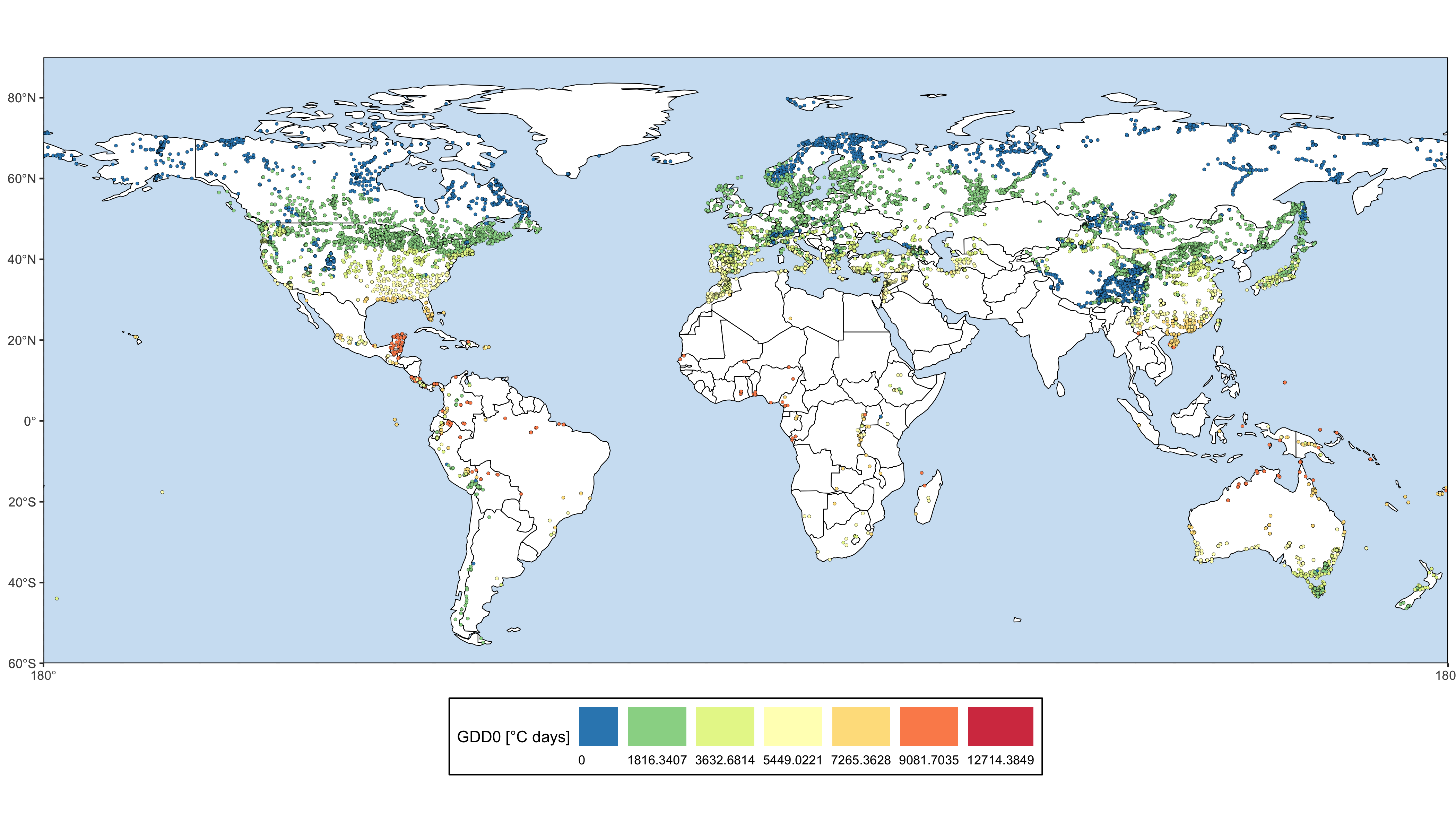
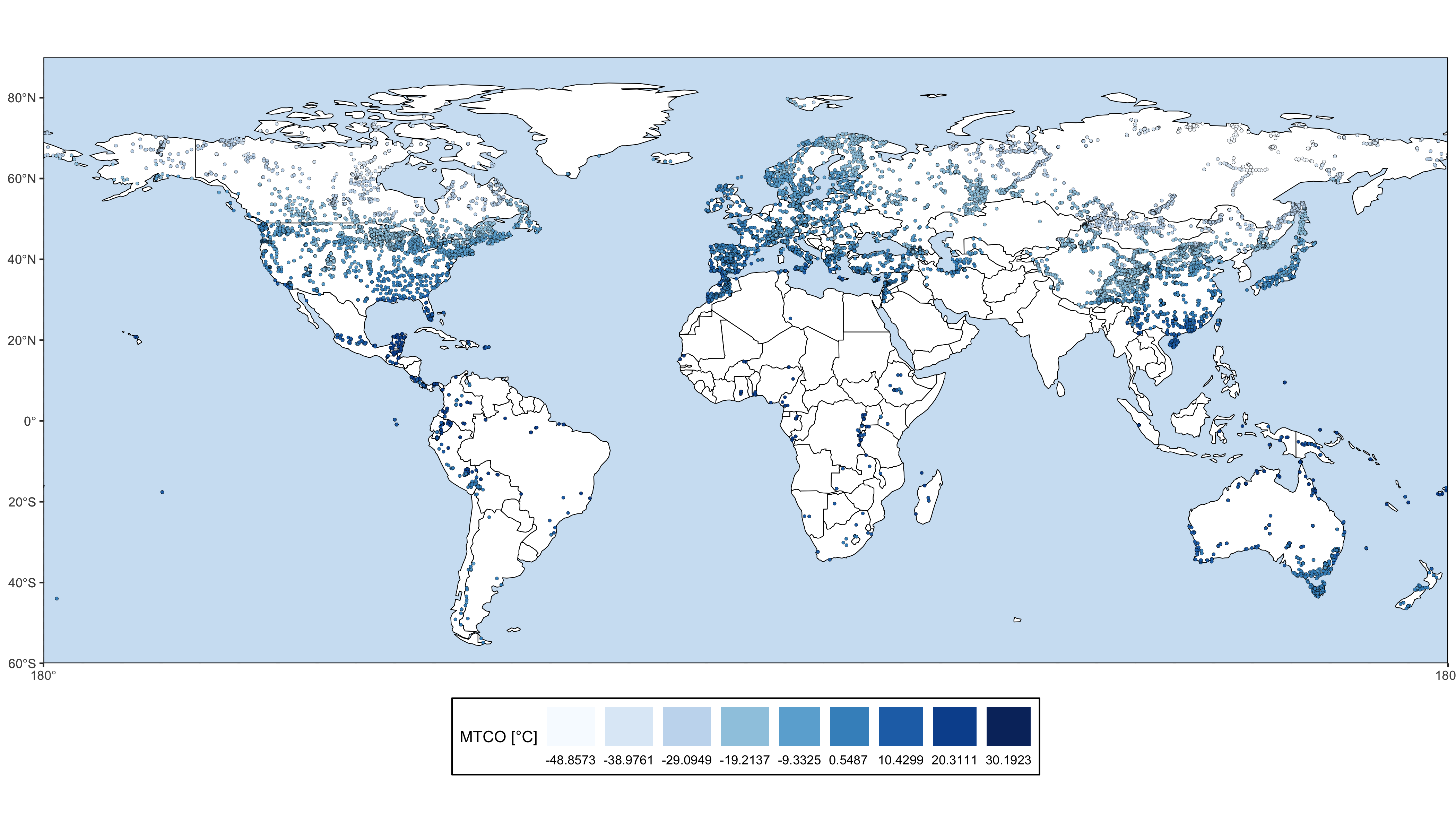
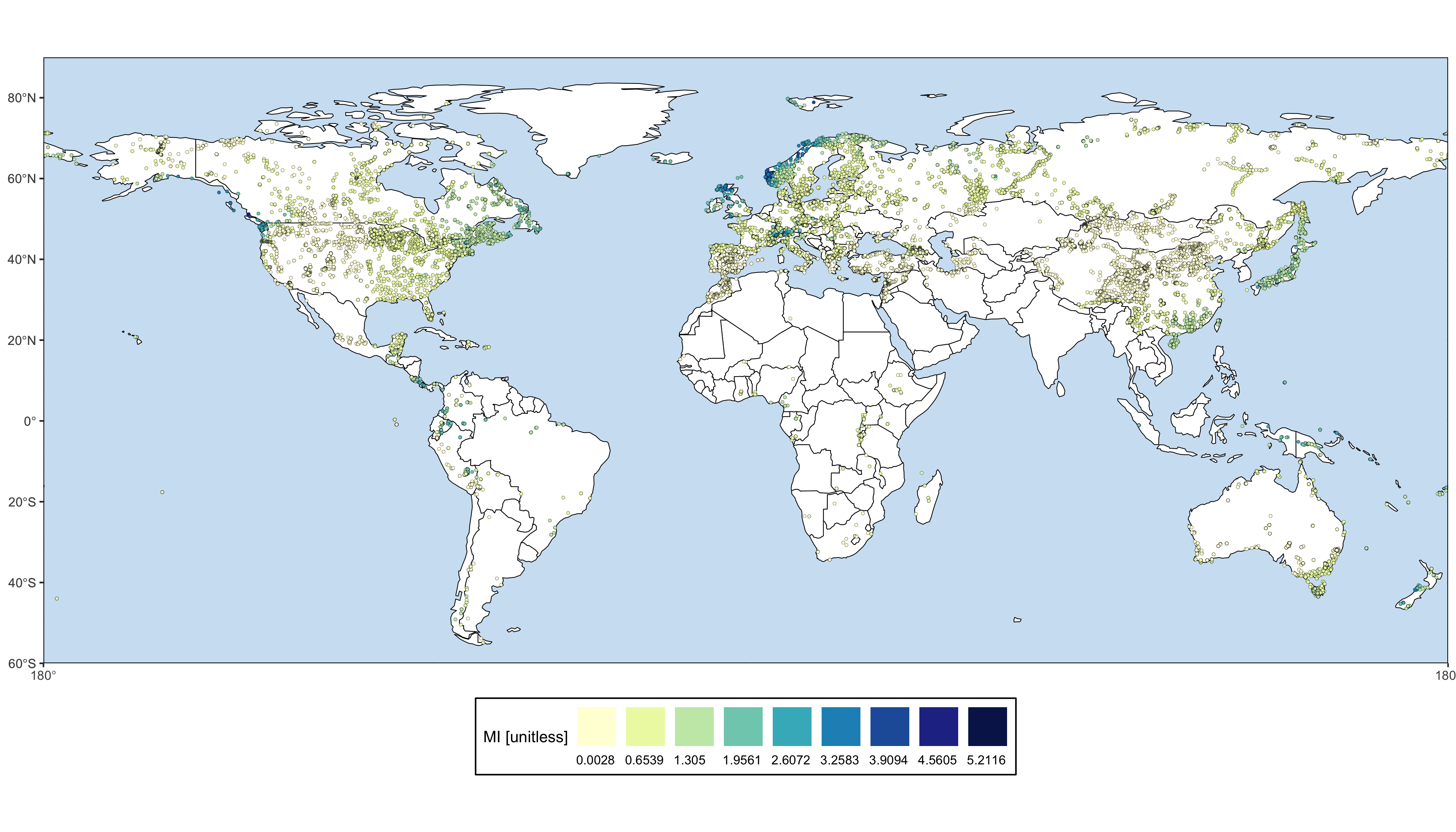
The goal of smpds is to provide access to the SPECIAL Modern Pollen Data Set for Climate Reconstructions (SMPDS).
Installation
You can(not) install the released version of SMPDS from CRAN with:
install.packages("smpds")And the development version from GitHub with:
# install.packages("remotes")
remotes::install_github("special-uor/smpds")Example
Create a snapshot of entities
The function smpds::snapshot takes few different parameters and based on the first one, x, it returns a variety of snapshots.
This function returns a list with 3 components:
-
entity: data frame (tibbleobject) with the metadata associated to the entities. -
climate: data frame (tibbleobject) with the climate and vegetation reconstructions. This one can be linked to theentitytable using the column calledID_SAMPLE. -
pollen_count: list of data frames (tibbleobjects) containing the pollen counts for 3 levels of “amalgamation”:cleanintermediateamalgamated
entitytable using the column calledID_SAMPLE.
⚠️ NOTE: the output is returned “invisibly”, so you should assign the output of the function to a variable.
output <- smpds::snapshot(...)
output$entity
output$climate
output$pollen_count$clean
output$pollen_count$intermediate
output$pollen_count$intermediateUsing the entity_name
smpds::snapshot("juodonys_core")
#> # A tibble: 1 × 6
#> ID_SITE ID_ENTITY ID_SAMPLE site_name entity_name pollen_counts$clean
#> <int> <int> <int> <chr> <chr> <int>
#> 1 3890 7901 1 Juodonys juodonys_core 1
#> # … with 2 more variables: pollen_counts$intermediate <int>, $amalgamated <int>Using the site_name
smpds::snapshot("Petresiunai", use_site_name = TRUE)
#> # A tibble: 1 × 6
#> ID_SITE ID_ENTITY ID_SAMPLE site_name entity_name pollen_counts$clean
#> <int> <int> <int> <chr> <chr> <int>
#> 1 6690 14229 2 Petresiunai petresiunai_121 1
#> # … with 2 more variables: pollen_counts$intermediate <int>, $amalgamated <int>Using the ID_ENTITY
smpds::snapshot(2)
#> # A tibble: 1 × 6
#> ID_SITE ID_ENTITY ID_SAMPLE site_name entity_name pollen_counts$clean
#> <int> <int> <int> <chr> <chr> <int>
#> 1 1 2 9710 05-Mo 05-Mo-10 1
#> # … with 2 more variables: pollen_counts$intermediate <int>, $amalgamated <int>Using the ID_SITE
smpds::snapshot(3, use_id_site = TRUE)
#> # A tibble: 1 × 6
#> ID_SITE ID_ENTITY ID_SAMPLE site_name entity_name pollen_counts$clean
#> <int> <int> <int> <chr> <chr> <int>
#> 1 3 37 15871 11 [HFL11] HFL11 1
#> # … with 2 more variables: pollen_counts$intermediate <int>, $amalgamated <int>Extracting multiple records at once
smpds::snapshot(1:10)
#> # A tibble: 10 × 6
#> ID_SITE ID_ENTITY ID_SAMPLE site_name entity_name pollen_counts$clean
#> <int> <int> <int> <chr> <chr> <int>
#> 1 1 1 9709 05-Mo 05-Mo 10
#> 2 1 2 9710 05-Mo 05-Mo-10 10
#> 3 1 3 9711 05-Mo 05-Mo-11 10
#> 4 1 4 9712 05-Mo 05-Mo-12 10
#> 5 1 5 9713 05-Mo 05-Mo-13 10
#> 6 1 6 9714 05-Mo 05-Mo-14 10
#> 7 1 7 9715 05-Mo 05-Mo-15 10
#> 8 1 8 9716 05-Mo 05-Mo-16 10
#> 9 1 9 9717 05-Mo 05-Mo-17 10
#> 10 1 10 9718 05-Mo 05-Mo-18 10
#> # … with 2 more variables: pollen_counts$intermediate <int>, $amalgamated <int>Extracting all the records at once
This will run ****very slow****, so if only few entities are required, it would be better to indicate which, based on the previous examples.
out <- smpds::snapshot()Export data as individual CSV files
The function smpds::write_csvs takes to parameters:
-
.data: a list of classsnapshot, this one can be generated using the functionsmpds::snapshot(see previous section). -
prefix: a prefix name to be included in each individual files, this prefix can include a relative or absolute path to a directory in the local machine.
Without a path
`%>%` <- smpds::`%>%`
smpds::snapshot("juodonys_core") %>%
smpds::write_csvs(prefix = "juodonys_core")
#> # A tibble: 1 × 6
#> ID_SITE ID_ENTITY ID_SAMPLE site_name entity_name pollen_counts$clean
#> <int> <int> <int> <chr> <chr> <int>
#> 1 3890 7901 1 Juodonys juodonys_core 1
#> # … with 2 more variables: pollen_counts$intermediate <int>, $amalgamated <int>Including a path
`%>%` <- smpds::`%>%`
smpds::snapshot("juodonys_core") %>%
smpds::write_csvs(prefix = "/special.uor/epd/juodonys_core")Code of Conduct
Please note that the SMPDS project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Publications
This package is a companion to the following dataset:
Villegas-Diaz, R., Harrison, S. P., 2022. The SPECIAL Modern Pollen Data Set for Climate Reconstructions, version 2 (SMPDSv2). University of Reading. Dataset. https://doi.org/10.17864/1947.000385